ZOOPLANKTON FORMULATIONS IN MARINE BIOGEOCHEMICAL MODELS
PI and organization: U. Daewel (HZG)
Co-Is: J. Schulz-Stellenfleth (HZG), C. Schrum (HZG), A. Samuelsen (NERSC), V. Çağlar Yumruktepe (NERSC)
The Project ZOOMBI set out to add spatially and temporally explicit representation of critical processes of the marine biogeochemical forecast. Thereby addressing major model uncertainties related to zooplankton parameterization and its role in the biological pump and, in the process, making the first steps towards the integration of higher trophic levels in the model. Therefore we identified two main processes that required additional attention and formulated our objectives accordingly:
Objective 1. Add a dynamical representation of zooplankton mortality by implementing a consistently formulated end-to-end (E2E) (from nutrients to fish) ecosystem model ECOSMO E2E simple enough to be applicable in operational services.
During the project both macrobenthos and two types of fish functional groups (dominantly zooplanktivores and dominantly pisci- and benthivores) were implemented in the model system TOPAZ-ECOSMO, which is currently used for the Arctic MFC. Knowing that fish in our study area performs active migration behaviour, we additionally implemented a migration scheme for the fish groups and successfully tested a simple migration rule where fish follows gradients of predator to prey ratios.
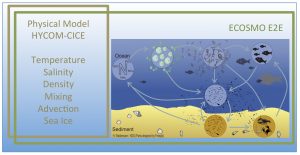
Figure 1. Simplified representation of the ECOSMO E2E model system and its coupling to the physical model HYCOM-CICE and the relevant environmental factors that need to be exchanged between models.
The ecosystem model benefits from these new developments in two ways. First, the representation of zooplankton grazing becomes dynamic and more process oriented. Second, the introduction of higher trophic level functional groups might potentially pave the way for new products on higher trophic level production CMEMS MFCs.
Objective 2. Improving the representation of sinking organic material in marine ecosystem models.
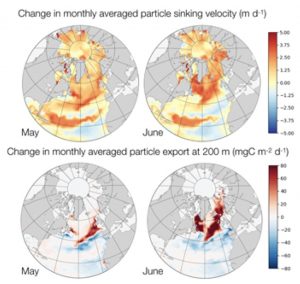
Figure 2. Simulated monthly average changes in particle sinking speed and export at 200 meters in a coarse North Atlantic and Arctic model domain.
Detritus sinking is one of the processes that, despite being complex and depending on the ecosystem functioning of a marine system, is often simplified to a large degree in marine ecosystem models. However, it has large effects on both nutrient recycling and carbon export. In ZOOMBI we implemented a novel formulation for detritus sinking velocity that treats detritus sinking as an additional state variable, which accumulates and contains information about the detritus composition and related changes in sinking speed. Using this we could show that the modelled organic matter export can be simulated more realistically in dependence of the ecosystem functioning, while adding only a minimum of computational extra costs to the model system.
Objective 3. Identify the potential of data assimilation for reducing uncertainties in the E2E ecosystem model using appropriate statistical methods.
In a final step we tested the potential for data assimilation to improve the newly implemented variables for fish. We could show that, when assuming hypothetical measurements for phytoplankton, data assimilation of the latter would not only improve the phytoplankton but also the new fish variable when an appropriate time lag is considered.